Drug discovery and development
We identify, develop and commercialise promising ideas into successful cancer therapeutics, diagnostics and enabling technologies.
The joint AstraZeneca–Cancer Research Horizons Functional Genomics Centre is collaborating with EMBL’s European Bioinformatics Institute to build its single-cell CRISPR capability. Find out how this technology advances drug discovery through genotype to phenotype correlation.

The emergence of CRISPR technologies has enabled rapid and precise gene editing opportunities. The Functional Genomics Centre (FGC) – a joint venture between Cancer Research Horizons and AstraZeneca – has been deploying CRISPR technology to advance cancer research. Located in Cambridge Biomedical Campus, the FGC comprises 28 cancer and computational scientists. To accelerate the discovery of new cancer medicines, the FGC aims to be the world’s leading centre of excellence in genetic screening, cancer modelling and big data processing.
The FGC uses pooled CRISPR screening to identify new targets for cancer drug discovery. Pooled CRISPR screens introduce a guide RNA (gRNA) library into a population of cells that can then turn genes off (knockout, CRISPRn; knockdown, CRISPRi or CRISPRoff) or turn genes on (activation, CRISPRa) depending on the type of CRISPR system. These libraries can be genome-wide or bespoke libraries of gene subsets depending on the biological question. The experimental conditions are optimised to ensure that each cell receives only a single gRNA, ensuring that only a single gene is perturbed in any one cell. These cells can then be subjected to a treatment (e.g., a drug) followed by an assessment of the change in guide composition by next-generation sequencing. For instance, gRNA distributions can be compared before and after a drug challenge. Depletion of specific gRNAs after drug treatment may indicate genes that sensitise cells to the drug and be a novel target to tackle drug resistance (Figure 1)1,2.
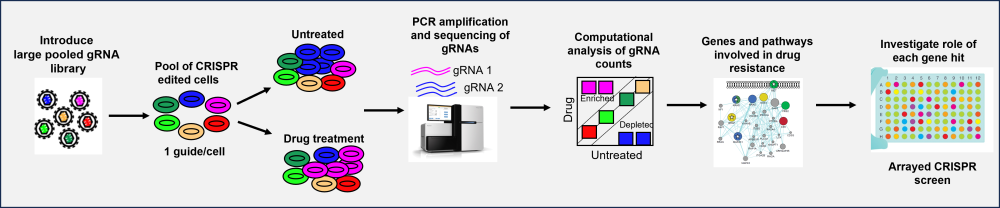
Pooled CRISPR screening enables unbiased target discovery at a genome-wide scale. However, the phenotypic readout of pooled screens is also averaged across a bulk of cells, with no indication about the changes occurring in individual cells to gain mechanistic insight3. This means that the hits identified by pooled CRISPR screens require individual validation and testing to elucidate the biological role of each gene hit of interest. A typical pooled screen can generate tens to hundreds of hits, many of which could be false positives, and it would require enormous resource to validate all the hits. Furthermore, pooled screens require an extraordinary number of cells, which makes them challenging to perform in finite biological models, such as patient material. Thus, it would be very beneficial to obtain rich multidimensional phenotypic information alongside the gene perturbation that can reveal the underlying molecular nature of phenotypes.
As such, pooled CRISPR screens can be combined with single-cell RNA sequencing, which enables the transcriptional response to be read out for each gRNA edit, across thousands of single cells. This instantly provides a detailed understanding of the biological role of each edited gene, enabling improved phenotype–genotype correlation. As a result, single-cell CRISPR screens significantly shorten the time for an initial validation of drug candidates and provide additional biological insights compared to the combination of pooled and arrayed screens (Figure 2)1. Common single-cell CRISPR techniques include cellular indexing of transcriptome and epitopes by sequencing (CITE-seq), CRISPR droplet sequencing (CROP-seq) and Perturb-seq4. Perturb-seq and CROP-seq predominantly use CRISPR interference (CRISPRi), where gene expression is blocked, in comparison to CRISPR knockout, where genes are rendered non-functional5. While cost currently prevents single-cell CRISPR screens at a genome-wide scale, these screens can be adopted as both the first screen (primary) to identify and validate the drug hits from a smaller predefined (bespoke) library, as well as the secondary screen to validate hits from a genome-wide pooled screen6.
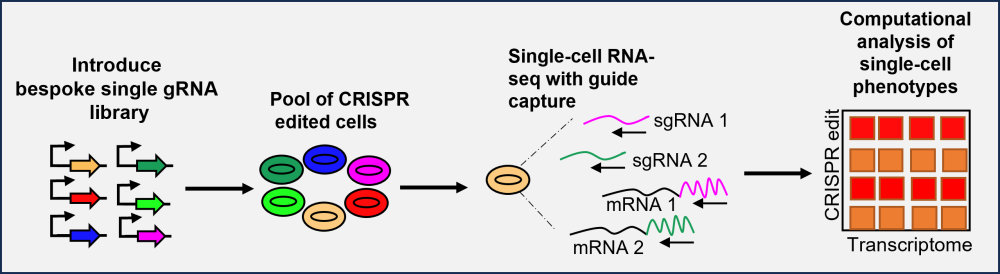
Excitingly, in the last year, the FGC initiated a collaboration with the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI) to deploy and improve Perturb-seq, a single-cell CRISPR capability. Fundamental to building a single-cell CRISPR capability at the FGC was the collaboration with Dr Magdalena Strauss, Sir Henry Wellcome fellow at the EMBL-EBI. Based on previous collaborative work at the EMBL-EBI and Sanger8, the FGC and EMBL-EBI together improved important elements of a standard single-cell analysis pipeline9. This enabled analysis of a complex dataset with both gene edits and transcriptomic information for every cell.
In this context, FGC conducted single-cell CRISPR experiments, not only attempting to replicate published data, but also advancing its applications. The results were successful, demonstrating good concordance to the published dataset. This has established a functional single-cell CRISPR screen pipeline for future AstraZeneca and Cancer Research Horizons projects at the FGC, as well as projects at the EMBL-EBI. Besides access to the single-cell CRISPR technology across three organisations, a key objective of this collaboration is to create a guidance document and build an analysis pipeline for performing single-cell CRISPR experiments, which will benefit the wider scientific community.
Pooled CRISPR screens require extensive data analysis, secondary screens, and a lengthy process to validate potential drug targets, whereas single-cell CRISPR screens provide direct genotype to phenotype correlation. A key benefit of having a single-cell CRISPR platform is the increased knowledge gained on potential drug targets, which increases the chances of finding the right target for drug discovery compared to standard pooled CRISPR screens. Furthermore, in single-cell CRISPR screens, we can use the transcriptomic readout for each gene knockout to determine which guides in a library are functional and thus can improve guide design for future guide libraries. Notably, through this collaboration, we have established a single-cell knockout (scRNA-CRISPRn) workflow, which has the added benefit of enabling us to use our existing and benchmarked CRISPR knockout libraries, saving further costs and analysis time.
With these efficiencies, we hope to accelerate the speed in which new medicines reach cancer patients. Ultimately, this single-cell CRISPR capability build demonstrates a highly successful partnership between academia and industry, setting a high standard for future collaborations. We hope this inspires an increasing number of cross-functional collaborations in the future.

Nikhil Gupta, Magdalena Strauss, Malwina Prater, Carlos Company, Khalid Saeed, Curtis Hart, Andy Sayer, Marica Gaspari, Alex Kalinka, Ultan McDermott, Gregory J Hannon, David Walter and Doug Ross-Thriepland