Functional Genomics Centre
The Cancer Research Horizons-AstraZeneca Functional Genomics Centre delivers pooled CRISPR screens to identify and validate new drug targets and...
The team at the Cancer Research Horizons–AstraZeneca Functional Genomics Centre generates vast amounts of data from their CRISPR screens every day. Here’s how they’re building a database to make sense of it all.
CRISPR-Cas9 technology has revolutionised genetic research and the way we discover new cancer medicines by offering unprecedented precision in editing DNA. CRISPR uses two key components, a Cas9 cutting enzyme and a guide RNA (gRNA molecule), which directs the cutting enzyme to the target sequence to be edited1. CRISPR holds promise for cancer research, as it enables scientists to identify genes important to cancer development and drug resistance. To harness the full potential of CRISPR against cancer, the Functional Genomics Centre (FGC) was established as a joint initiative between AstraZeneca and Cancer Research Horizons to achieve this mission.
In previous articles, we have discussed how CRISPR improves the efficiency of drug discovery (Using CRISPR to accelerate drug discovery) and the emerging technologies we are establishing (Building a single-cell CRISPR capability). Now, we talk data – how we analyse, interpret and find understanding in the vast amounts of information generated every day. The FGC primarily runs pooled CRISPR screens, where a gRNA library, containing hundreds to thousands of gRNA molecules, targets thousands of genes for editing. We run pooled CRISPR screens both Cancer Research Horizons and AstraZeneca project collaborators and a future collaborative goal aims to combine data from both organisations and establish a robust quality control (QC) system. This will ensure the data is accurate and reliable, therefore giving more confidence in the genes identified as potential new cancer drug targets.
We spoke to members of the teams responsible to answer the key questions about building the FGC’s CRISPR database. Thank you to Diego Fontecilla (Infrastructure Engineer, Cancer Research Horizons), Yasantha Soysa (Database Engineer, AstraZeneca), Alex Kalinka (Principal Bioinformatician, Cancer Research Horizons), Daniel Barrell (Director, Bioinformatics, AstraZeneca), and David Leach (Lead Solution Architect, Cancer Research Horizons) for the valuable discussions.

CRISPR screens involve systematically disrupting thousands of genes within cancer cells to pinpoint those essential for survival or drug response. This generates a wealth of data that must be analysed. The process begins with extracting and sequencing DNA from cell pellets taken from the lab at different timepoints and conditions in a CRISPR screen. Bioinformaticians then use the pipeline to compare specific regions of DNA sequences, also known as genes, across all the different samples. From this, the pipeline impressively outputs a hit list of genes that are enriched or depleted between different conditions tested in a project. We then curate and incorporate these gene "hits" into a CRISPR database, creating a comprehensive repository of genetic vulnerabilities in cancer.
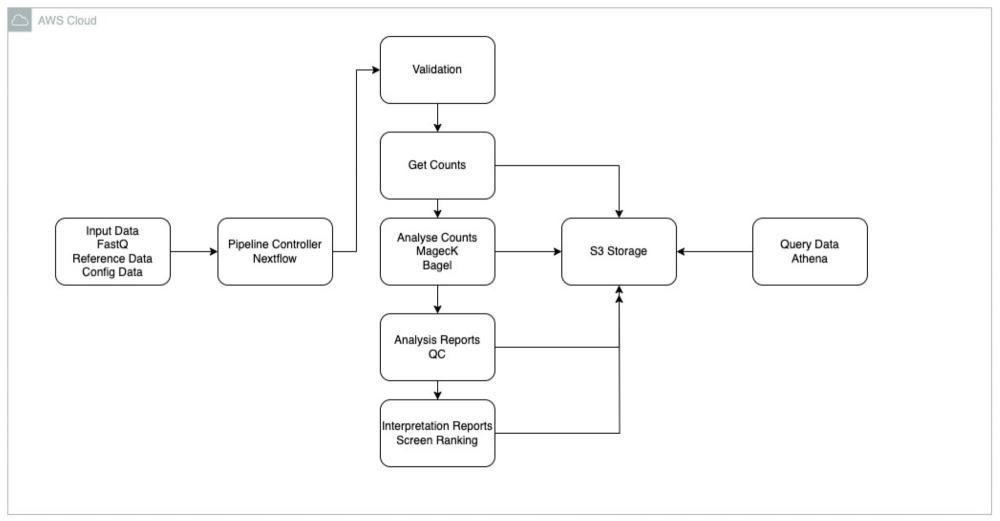
The CRISPR database combines CRISPR screen data from all FGC projects into one, easy to access place. Currently, both AstraZeneca and Cancer Research Horizons have been developing their own CRISPR databases in parallel, capturing data from either AstraZeneca- or Cancer Research Horizons-sponsored FGC projects separately. We are now combining both organisations’ independent databases to create a joint QC CRISPR database for all FGC projects.
The AstraZeneca database contains data from 29 FGC projects. This is a culmination of 281 screens, 83 cell lines and 51 treatments. We decided at an early stage to include data from both pooled CRISPR screens and arrayed CRISPR screens. Scientists typically perform arrayed screens after pooled screens, where they edit each individual gene separately1. This adds richer data on the role of individual gene functions and aims to help predict how likely a hit will validate in future downstream work. AstraZeneca also captures over 1,000 public human screens from the BioGRID Open Repository of CRISPR screens (ORCS)2.
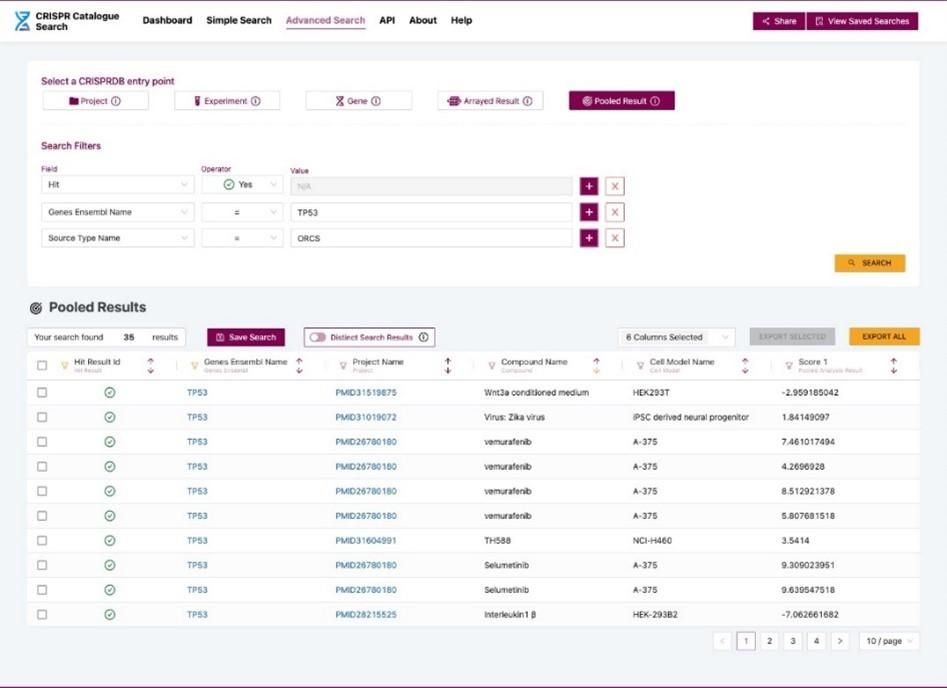
AstraZeneca’s database is built around two main CRISPR screen algorithms: (1) MAGeCK, a method for prioritising each gRNA molecule based on gRNA counts under different conditions, and the genes and pathways those gRNAs encode3; and (2) BAGEL, a computational framework for identifying essential genes4. These systems create model outputs and normalise CRISPR screen data.
Comparatively, the current Cancer Research Horizons internal database contains information from approximately 600 samples across 30 projects and 117 CRISPR screens. Users can make comparisons between cell lines, timepoints and gRNA count data, as well as any desired statistics. Unlike the AstraZeneca database, the Cancer Research Horizons equivalent contains all gRNA count data, in addition to the CRISPR screen algorithms, MAGeCK and BAGEL, which enables robust QC monitoring across project and over time.
Having a database provides the ability to search a vast amount of data quickly and identify trends. Within each organisation, the user can select a gene of interest and view all the CRISPR screens in which this gene was identified as a hit and in which cell model. A database also enables you to compare commonly enriched and depleted hits across different internal projects, particularly useful for analysing drug-gene CRISPR screens. This information can be valuable for determining a gene’s biological relevance and understanding more about the pathways a gene is involved in.
In addition, the database can provide QC metrics regarding screen performance. It is rare for a CRISPR screen to fail, but for those screens that exhibit a lower performance, identifying how the signal to noise ratio affects the number of hits is crucial. We typically observe lower performance in screens where Cas9 has been delivered into cells by electroporation compared to lentiviral transduction. Having the ability to identify these anomalies is important in making changes to reduce noise and maximise hit quality in future.
The Cancer Research Horizons database has made fantastic use of data on gRNA counts, which indicate the abundance of each gRNA molecule and evaluate how well the gRNA library has edited and so performed in a CRISPR screen. Besides targeting genes of interest, gRNA libraries have non-targeting gRNAs – control gRNA molecules that intentionally do not target genes for editing. As there is no disadvantage to having a non-targeting gRNA in the library, these gRNAs often slightly enrich in abundance over time in a screen and can be used as a metric for assessing gRNA library performance5.
Interestingly, the Cancer Research Horizons database showed that several non-targeting control gRNA molecules in the FGC’s dominant library behaved abnormally, depleting significantly over time, as if they were targeting genes essential to cancer’s survival. The database even identified one non-targeting gRNA that had an off-target edit at a cancer-relevant gene, which causes death of cancer cells. As a result of this data, we have improved our gRNA library, selecting only non-targeting gRNAs that work with high confidence. This aims to reduce the chances of false positive drug targets being identified from our CRISPR screen data and prioritise the right targets for further validation.
For AstraZeneca, we have learnt massively from building a user interface website, which enables people to input queries into the database without knowledge of any database language. This has meant that for the first time, we have aggregated data across FGC projects. This has surprisingly made it easier to spot inconsistencies, such as in naming of drug compounds. With our database design being highly flexible, we aim to standardise all fields, like compound naming and fix prior data entries. This will ensure that users find all relevant entries when they search for them in the database.
The fact that both AstraZeneca and Cancer Research Horizons have been building their own databases in parallel has accelerated improvements. Sharing experiences and learnings of what works and what does not has been highly valuable in ensuring the best platform possible was created. AstraZeneca has a platform team who collaboratively share updates on database structure to Cancer Research Horizons builders and vice versa. For the future, this should enable Cancer Research Horizons to build a user interface like AstraZeneca did, but much more efficiently, using learnings and existing knowledge.
We also aim to co-develop new tools that will help us interpret and understand data from CRISPR screens. For example, we have developed a tool that allows us to use visual protein–protein networks based on the STRING database and layer on top different prior knowledge, such as how often a gene is mutated in cancer. These tools make it easier for any scientist to identify patterns and biological pathways, which ultimately lead to new hypotheses and targeted medicines6.
For Cancer Research Horizons, we are currently on our second version of the database. Our first database was far too complex, having to scan up to 2 billion rows of data to solve any given problem. Simplifying the design was therefore critical in ensuring an improved performance in our second version.
Now, Cancer Research Horizons’ database is built using Athena, the Amazon Web Services (AWS) database engine7, which operates on a self-service system. This means that the underlying database consists of multiple files that can be treated either as separate databases or combined into one database. In future, this gives Cancer Research Horizons the option to look at integrating publicly available databases, such as the Cancer Cell Line Encyclopedia (CCLE)8, into the CRISPR database. CCLE contains genomic, proteomic, drug response, epigenetic and transcriptomic datasets across around a thousand cancer cell lines and thus could add a huge resource of biological information onto the cell lines used in CRISPR screens in the database8.
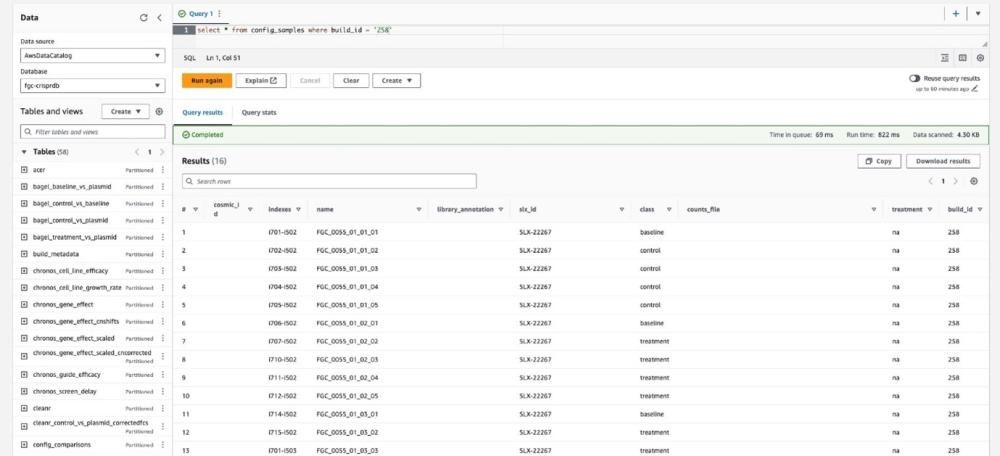
Cancer Research Horizons’ current goal for 2024 is to build a central CRISPR database combining the QC data from both its and AstraZeneca’s CRISPR screen projects. Currently, we drop QC report files into a project’s SharePoint location and if we discover an additional QC metric that we want to add to the report, we have to generate the analysis and report all over again. However, having a central QC database offers increased flexibility, in which users can calculate new QC metrics retrospectively for prior projects. This means that as we continue to improve our QC analysis process, we can gain more insightful data from ongoing and previous projects.
AstraZeneca’s current challenge in building the central CRISPR database is bringing all the QC data together, without sharing confidential information from each organisation. We have realised that even simple fields, such as sample labels, need more standardisation in naming than initially expected. However, once we overcome this problem, we envision the CRISPR database to be a tremendous resource for the CRISPR and cancer biology research field. It will be the first time that CRISPR QC data will be available across a plethora of essentiality and drug–gene interaction CRISPR screens, as well as a diversity of cancer models from cancer cell lines to immune cells and 3D models.
Our long-term vision is to make the central CRISPR QC database publicly accessible, by releasing it in a future publication. This would enable researchers to view which parameters improve or worsen screen performance and therefore improve the design and performance of future CRISPR screens. With improved CRISPR screen quality, this should increase the chances of identifying true positive genes as drug targets in the first stage of drug discovery, drug target identification.
The FGC was set up as a joint endeavour in 2019 to establish a world-leading centre for CRISPR screening that would make a difference to how we discover and develop new cancer medicines. This revolution is only possible if we can successfully analyse, interpret and make accessible the vast amounts of information generated by every screen. This article celebrates the great work the IT teams are doing across organisations and geographies to create the infrastructure and tools to help us achieve this mission. We need this diverse expertise to push the boundaries of science and advance how we develop medicines.
Alex Kalinka, David Leach, Diego Fontecilla, Chris Wood, Annie Kay, Shannon Ray and David Walter (Cancer Research Horizons). Daniel Barrell, Yasantha Soysa, Venkatraman Chandrasekaran, Biswa Ranjan Roul, Uka Osim, Mohammed Matin, Frida Moberg, Manasa Surakala, Neli Atanassova and Doug Ross-Thriepland (AstraZeneca).